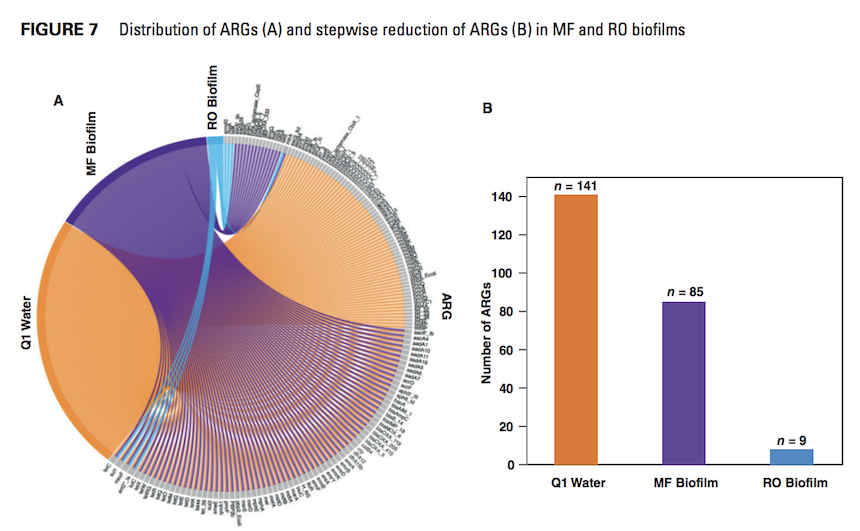
Part A shows a chord diagram listing ARG sequences in Q1 water and MF and RO biofilm samples. All ARGs are represented as a gray edge. A line connects each gene to each sample where it was found to be present. All the ARG sequences compared represent partial to complete gene coverage. Part B shows stepwise reduction of ARGs identified in MF and RO biofilms and undisinfected influent Q1. Four genes were found in common among all sample types
Culture-based methods identify only those organisms that can be grown, including pathogens; such methods underestimate and provide limited understanding of microbial communities. Polymerase chain reaction methods are rapid but limited. Deoxyribonucleic acid (DNA) sequencing technologies provide a method for identifying and characterizing the total microbial community in water by using next-generation sequencing (NGS) coupled with metagenomics analysis. In this study, NGS was used to identify and characterize the microbial community comprising fungi, protozoa, bacteria, pathogens, viruses, and antibiotic resistance genes (ARGs) in biofilms of the feed sides of microfiltration (MF) and reverse osmosis (RO) membranes in an advanced water purification facility that produces potable-quality water. The undisinfected influent to the treatment train was also characterized by NGS, providing the capability to monitor the microbial community of the secondary treated wastewater. This study represents the first application of whole genome shotgun sequencing of the entire microbial DNA by metagenomics to identify and characterize microbial communities in water and biofilms.
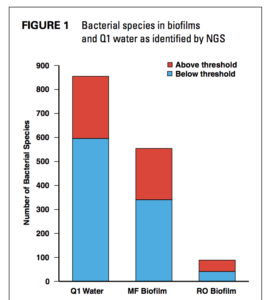
Q1—undisinfected influent, RO—reverse osmosis
Organisms above the threshold are identified with ≥95% specificity
determined by data-mining algorithm. Those identified as below the
threshold require further validation to confirm the identification.
Fungi, protozoa, and bacteria, and their ARGs were identified in undisinfected influent and in the MF biofilm, demonstrating reduction after MF treatment, particularly for pathogens and ARGs. Bacteriophages, viruses, and protozoa were detected in the influent water but not within the RO biofilm. Their absence after treatment demonstrated MF membrane treatment efficacy. Figure 1 summarizes the total bacterial species identified and their decrease during MF and RO treatment. DNA from microbial species considered to be opportunistic pathogens was detected in all samples with a decrease in species diversity. However, Citrobacter, Streptococcus spp., Bacteroidales, and Enterococci spp., which are indicator bacteria commonly identified in domestic sewage, were not detected in the RO biofilm but were identified in the undisinfected influent and the MF biofilm. These findings demonstrate that NGS can be used to evaluate advanced water treatment processes for microbial removal effectiveness.
Moreover, NGS can simultaneously characterize the microbial community, including the ARG population, in complex water matrixes. Studies are underway to develop simultaneous DNA and ribonucleic acid characterization. This will greatly expand the potential applications of NGS technology, including its use for viability assessments of the component species of microbial communities during water treatment. This study demonstrates a radical change and potential comprehensive approach to microbial water quality monitoring and treatment performance assessment. The improved ability to assess treatment performance will help ensure the safety of reused water as well as water systems in general.
This feature is a summary by corresponding author, Menu B. Leddy, a principal scientist at Orange County Water District. For full text: pdf
